Help for using ZincExplorer
The protein sequence input to our server should be in RAW or FASTA format. Only the 20 conventional amino acid symbols are supported. The sequence should contain 15 or more residues and should contain at least one CHED (i.e. CYS, HIS, GLU and ASP). The email address is required since the result will be sent to it.
The prediction output consists of four items:
- Position
- Score
- Zinc-binding site annotations (YES for zinc-binding sites, NO for non-zinc-binding sites).
- Interdependent relationship
Three measurements, i.e. Precision, Recall and AURPC were used to evaluate the prediction performance.
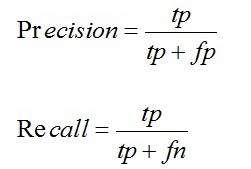
tp, fp, fn and tn denote true positives, false positives, false negatives and true negatives.
We will email you the result when the job is done. You can also query your prediction result in the job list page or input you job ID in the query form.