PPIRA is a database that contains protein-protein interactions between R. solanacearum and A. thaliana, which were predicted by two bioinformatic methods, the interolog and domain-based methods. When you query a R. solanacearum protein, a table (e.g. Table 1) will be returned. This table contains A. thaliana proteins which may interact with the queried R. solanacearum protein. Similarly, if you query an A. thaliana protein, a table containing R. solanacearum proteins which may interact with the queried A. thaliana protein will also be returned (e.g. Table 2). In the returned table, "ID" is the potential interacting partner of the queried protein. "Synonym" shows other types of the protein ID. "GO annotation" is the function annotation of the protein, which could be categorized into three parts, biological process, cellular component and molecular function. "Template in DIP" represents interacting protein pairs in the DIP database, from which the potential PPI was inferred. "Template in iPfam" denotes interacting domain pairs in the iPfam database, from which the potential PPI was inferred. These two columns show the numbers of templates available from the interolog and domain-based methods, respectively. From the returned tables, you can link TAIR, NCBI, genome annotations of R.solancearum in INRA, and the website of Gene Ontology to check the information of proteins.
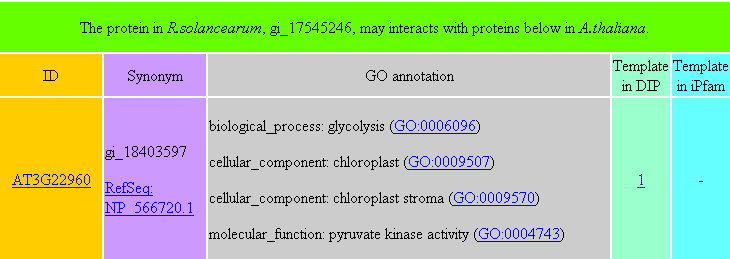
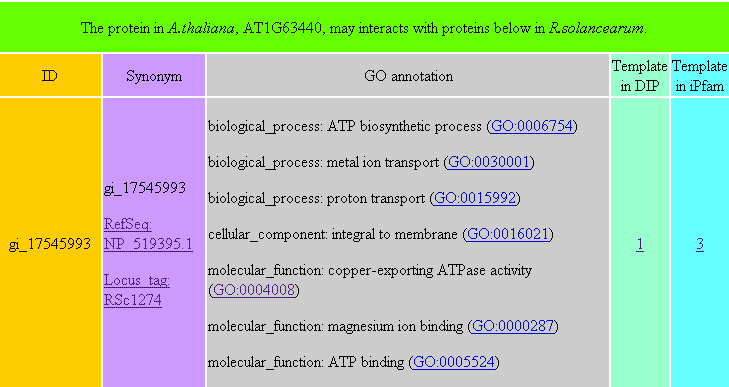
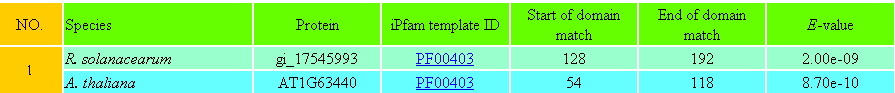
Detailed template information of each potential PPI is also provided (e.g. Table 3 and Table 4). In Table 3, the last four columns are four resulting parameters generated from the BLAST search. In Table 4, the last three columns are three resulting parameters generated from the Pfam search.
 |
 |