
Wiki
Type-III effectors are virulence proteins secreted by pathogenic bacteria to sabotage their host defense system. Because they are not only the powerful weapons of pathogens, but also probes for researchers to investigate the principle of host immunity, their functions have been intensively studied in recent years.Here, we gives a brief overview on type-III effectors and the principle behind our prediction algorithm, to help our users explain the results more rationally.
Type-III secretion system and type-III effector
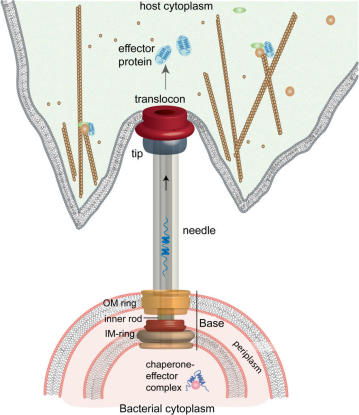
Type-III secretion system is a protein appendage found in several Gram-negative bacteria.
When the extracellular surface receptors of the host recognize the pathogen-associated molecular patterns(PAMPs), (PAMP-triggered immunity)PTI will be initiated to prevent the invasion of bacteria. However, some pathogenic bacteria have evolved a needle-like system to secreted effectors that help pathogen to survive and to escape an immune response. In response, plant resistance proteins sense effectors to activate effector-triggered immunity(ETI).
Most effectors in the host cells exert their function by mimicking the function of some proteins in the host. More information about type-III secretion system is abilable at http://en.wikipedia.org/wiki/Type_three_secretion_system.
Workflow of BEAN 2.0
BEAN 2.0
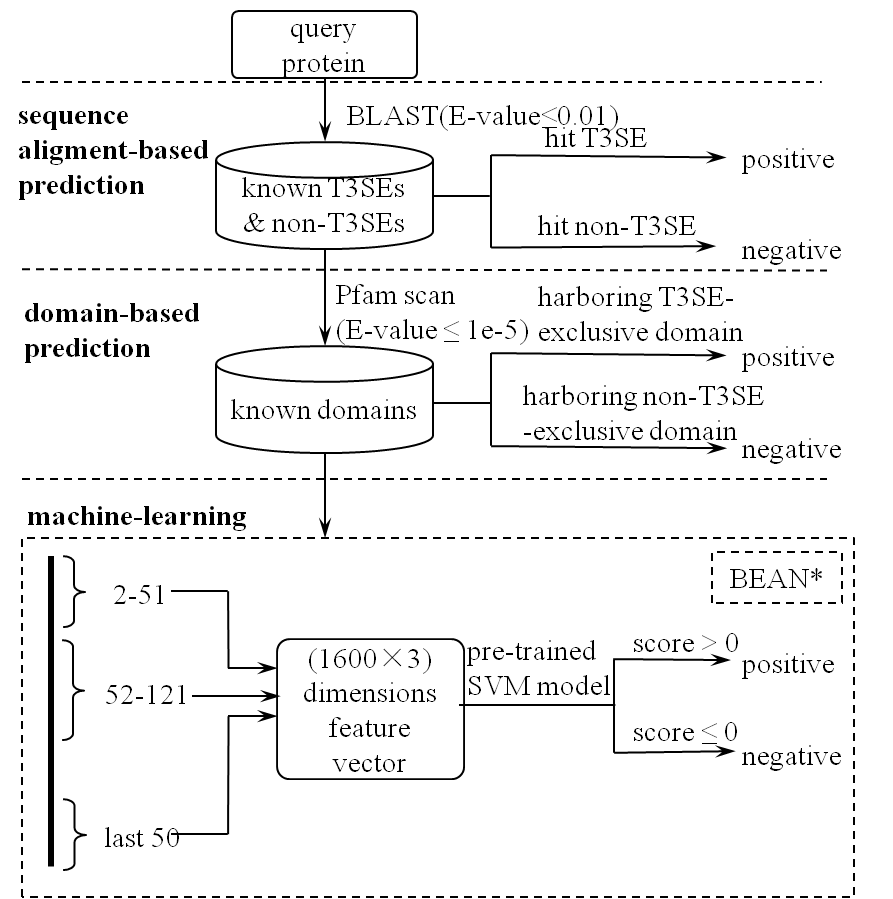
BEAN*
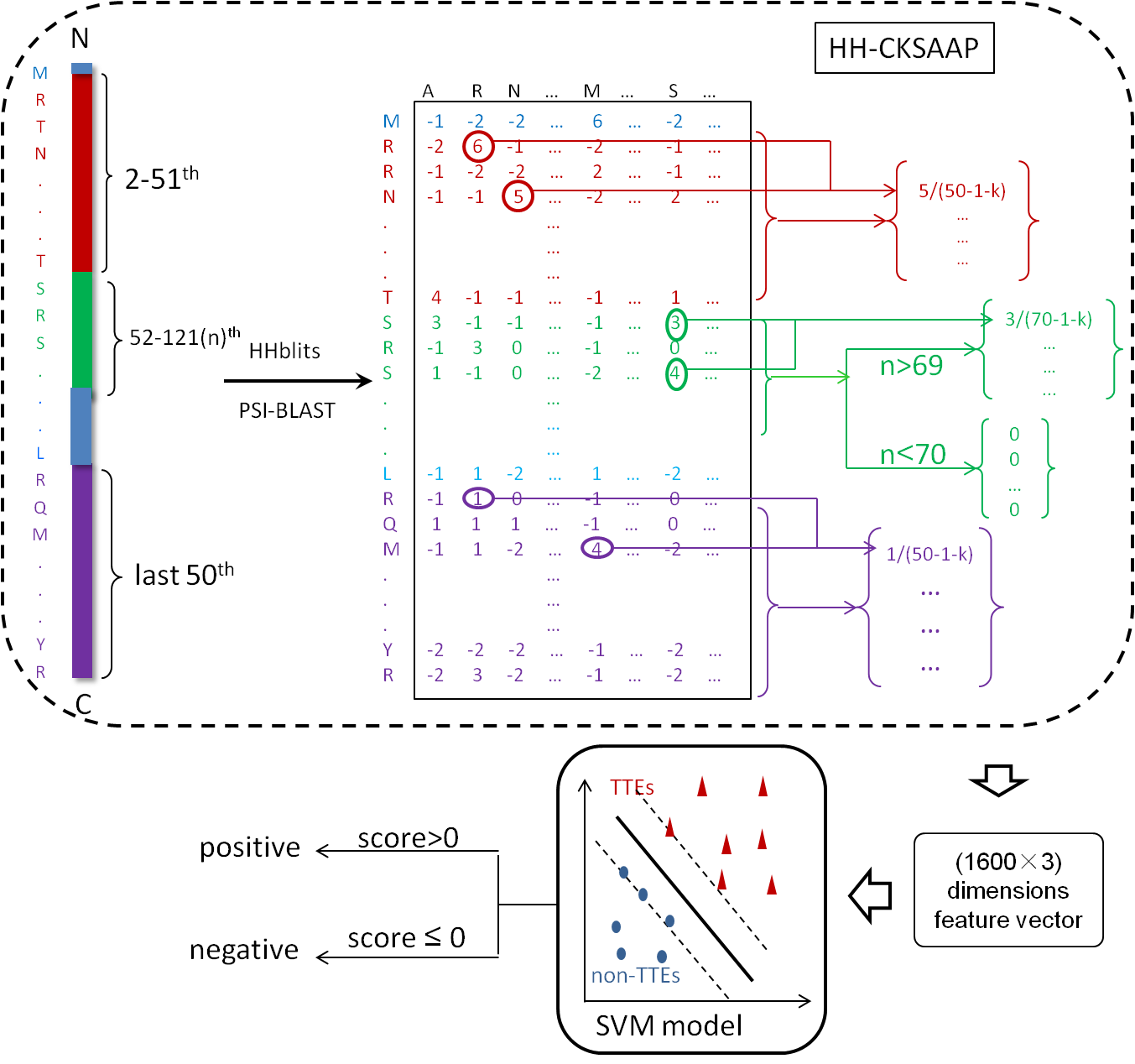
Alogrithm
Training data
We used 243 type-III effectors and 486 non-type-III effectors to train BEAN 2.0's classification model.
Encoding Methods
First, the training set is taken as the BLAST database, and each of the query proteins search against this database. Among all the hits , we consider the top hit(the minimum e-value) as the most valuable result. If the hit is corresponded to a effector then the query protein is directly assigned as a effector. Similarly if the hit is corresponded to a non-effector then the query protein is directly assigned as a non-effector.
There are also no hits for many proteins, then the HMM-based sequence search will be done using the Pfam database. Each protein in our training set is searched against the Pfam database using the HMM search at an e-value threshold of 1e-5. The domains of query proteins are searched against these two databases.
Database 1: Exclusively effector domains occurring only in effectors
Database 2 :Exclusively non-effector domains occurring only in non-effectors
A protein was assigned as a effector if it has an exclusive effector domain and was assigned as a non-effector if it has an exclusive non-effector domain.
If the domain is not contained in these databases, BEAN* will be used for prediction. In BEAN* , we divide a protein into three parts: 2~51 amino acids of the N terminus, 52~121 amino acid of the N terminus, 50 amino acids of the C terminus. The N terminus and C terminus are denoted by a 3200-dimension vector. If the number of the amino acids in the middle part is larger than 69, a 1600-dimension vector using the the same algorithm in BEAN would be generated. Otherwise, a 1600-dimension vector using all of the score equal "0" would be used to denoted the remaining part. Then a 4800-dimension vector generating from HH-CKSAAP is used to denote a protein. The 4800-dimension feature vectors are used to train a SVM model.
Machine learning technology
We use a machine learning technology named Support Vector Machine (SVM) to build BEAN. As a state-of-the-art machine learning technology, SVM has been widely used in such as handwriting recognition, protein functional site prediction, financial analysis and computer vision etc. More information about SVM can be found at SVM.org.
Instructions
For some instructions of the website, please turn to "help" page or download Userguide.pdf.
Publications
(1) Dong, X., et al. (2013). "Using weakly conserved motifs hidden in secretion signals to identify type-III effectors from bacterial pathogen genomes." PLoS One 8(2): e56632.
(2)Dong, X., et al. (2015). "BEAN 2.0: an integrated web resource for the identification and functional analysis of type III secreted effectors."Database (Oxford)
Reference
[1] Chatterjee, S., et al. (2013). "Structure and biophysics of type III secretion in bacteria." Biochemistry 52(15): 2508-2517.