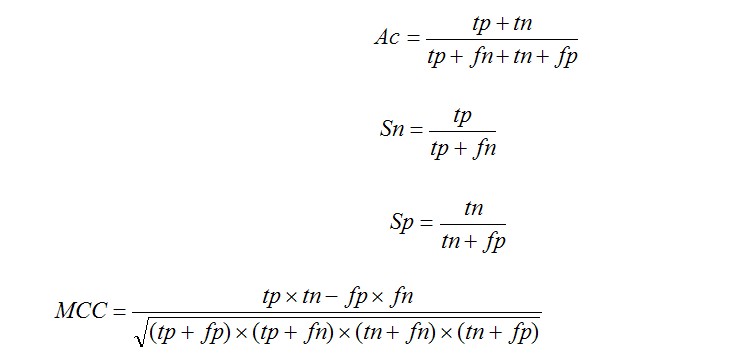About CKSAAP_UbSite
CKSAAP_UbSite is a web server that could predict ubiquitination sites in proteins. With the assistance of SVM, the highlight of CKSAAP_UbSite is to employ the composition of k-spaced amino acid pairs as input feature vector. It was trained and tested on a set of experimentally verified ubiquitination sites obtained from Radivojac et al (Proteins, 2010, 78: 365-380). The class-balanced accuracy and MCC of CKSAAP_UbSite reached 73.40% and 0.4694, respectively. Since Radivojac et al’s dataset was selected from the proteome of S. cerevisiae, the application of CKSAAP_UbSite should be favorable in the proteome of S. cerevisiae.
About hCKSAAP_UbSite 
hCKSAAP_UbSite(LATEST UPDATE): Our recently updated human ubiquitination site predictor hCKSAAP_UbSite is now available! To use this predictor, please select "H.Sapiens" in the organism list. This predictor was trained on a comprehensive human ubiquitination site dataset which is available at the dataset download page. Four SVM classifiers utilizing different amino acid pattern and propensities encodings were integrated via logistic regression. The resulting predictor, hCKSAAP_UbSite, achieved an AUC of 0.769 in 5-fold cross validation test and an AUC of 0.757 in an independent test.
Stand-alone program for hCKSAAP_UbSite is now available here.
Copyright
This server is free available to any user, but without any warranty.
For using CKSAAP_UbSite, Please cite:
Chen Z, Chen Y-Z, Wang X-F, Wang C, Yan R-X and Zhang Z (2011) Prediction of Ubiquitination Sites by Using the Composition of k-Spaced Amino Acid Pairs. PLoS ONE,6: e22930.
Chen Z, Zhou Y, Song J and Zhang Z (2013) hCKSAAP_UbSite: Improved prediction of human ubiquitination site by exploiting amino acid pattern and properties. BBA - Proteins and Proteomics, 1834: 1461-1467.